SCENIC
SCENIC is a computational method to deduce gene regulatory networks.
The main progress has three parts.
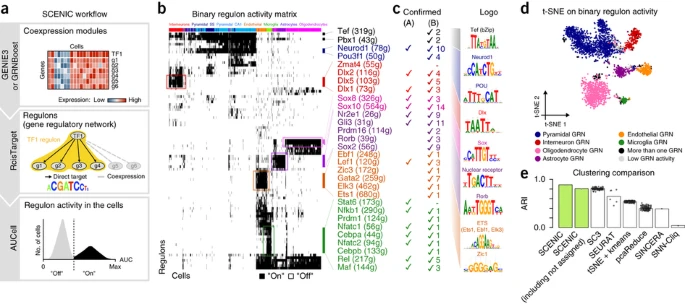
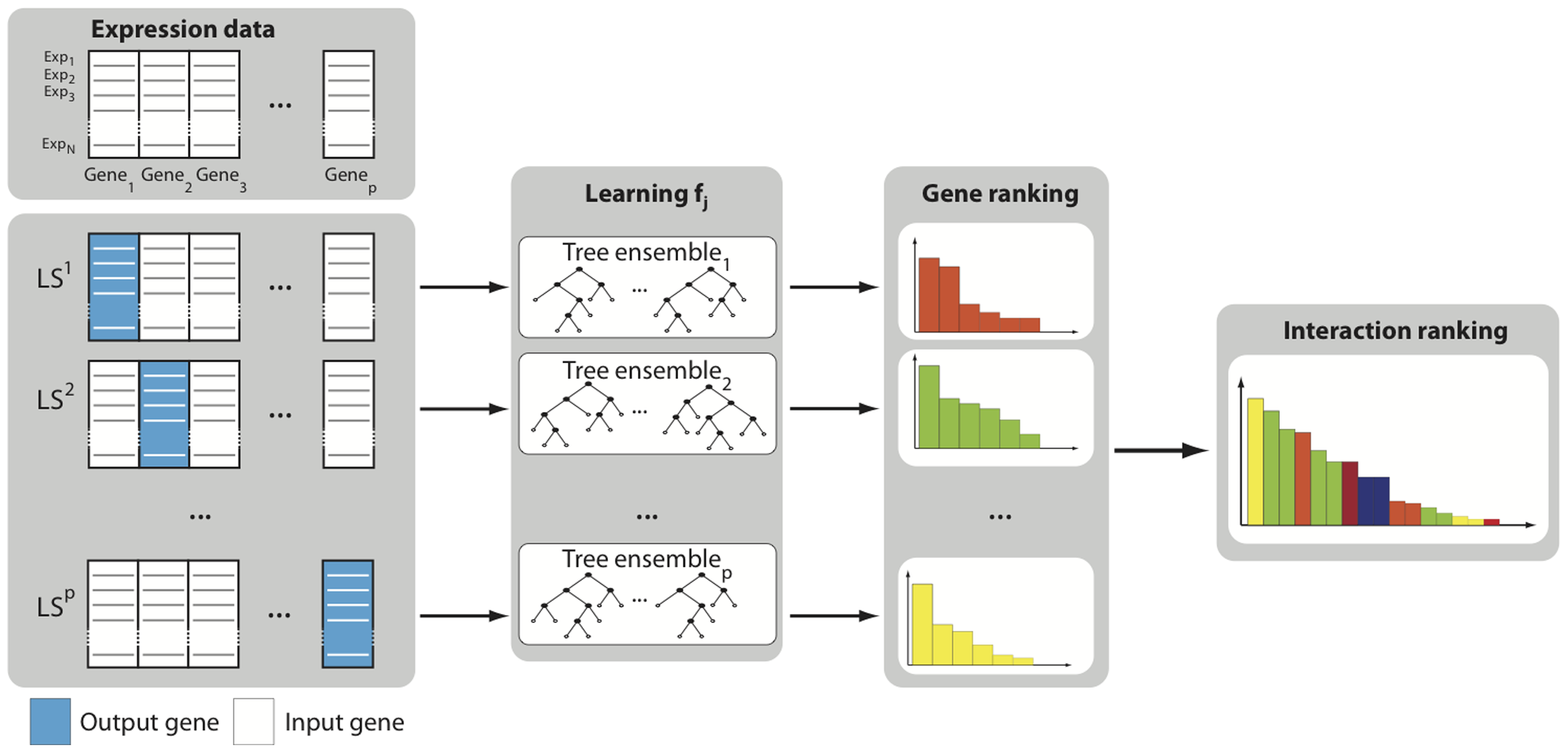
Part1. GENIE3
Through GENIE3 to identify genesets coexpressed with TF genes.
The method assumes that a specific gene expression depends on the other genes' expression in the network.
Denoting by \(x_k^{j-1}\) the vector containing the expression values in the \(k_{th}\) the experiment of all genes except gene j:
\(\mathbf{x}_k^{-j}=\left(x_k^1, \ldots, x_k^{j-1}, x_k^{j+1}, \ldots, x_k^p\right)^{\mathrm{T}}\)
Then,
\(x_k^j=f_j\left(\mathbf{x}_k^{-j}\right)+\varepsilon_k, \forall k\) , where \(\varepsilon_k\) is a random noise with zero means (conditionally to \(\mathbf{x}_k^{-j}\))
For j = 1 to p:
- Generate the learning sample of input-output pairs for gene j:
\(L S^j=\left\{\left(\mathbf{x}_k^{-j}, x_k^j\right), k=1, \ldots, N\right\}\)
Use a feature selection technique on \(L S^j\) to compute confidence levels \(w_{i, j}, \forall i \neq j\), for all genes except gene j itself.
Aggregate the p individual gene rankings to get a global scale of all regulatory links.
Part2. RcisTarget
RcisTarget identifies enriched TF-binding motifs and candidate transcription factors for a gene list.
In these two steps:
Step 1 selects DNA motifs significantly over-represented in the surroundings of the transcription start site (TSS) of the genes in the gene set.
Step 2, RcisTarget predicts candidate target genes (i.e., genes in the gene set that are ranked above the leading edge), like enrichment analysis.
Part3. AUCell
The input of AUCell is a regulon that contains TF and regulated genes.
AUCell calculates the enrichment of the regulon as an area under the recovery curve (AUC) across the ranking of all genes in a particular cell, whereby genes are ranked by their expression value.
Reference
- [1] SCENIC
- [2] GENIE3
- [3] RcisTarget